---
title: "Week 10 figures - Lectures 17 and 18"
editor: source
freeze: auto
execute:
message: false
warning: false
format:
html:
code-fold: true
author:
- name: An Bui
url: https://an-bui.com/
affiliation: UC Santa Barbara, Ecology, Evolution, and Marine Biology
affiliation-url: https://www.eemb.ucsb.edu/
published-title: "Lecture date"
date: 2024-06-03
date-modified: last-modified
categories: []
citation:
url: https://spring-2024.envs-193ds.com/lecture/lecture_week-10.html
---
```{r}
# cleaning
library(tidyverse)
# visualization
theme_set(theme_classic() +
theme(panel.grid = element_blank(),
axis.text = element_text(size = 18),
axis.title = element_text(size = 18),
text = element_text(family = "Lato")))
library(patchwork)
library(ggeffects)
library(flextable)
library(GGally)
library(equatiomatic)
# data
library(palmerpenguins)
# analysis
library(car)
library(performance)
library(broom)
library(DHARMa)
library(MuMIn)
library(lmtest)
```
# math notation
## simple linear regression
$$
E[y_i] = a + bx_i
$$
$$
var[y_i] = s^2
$$
## generalized form:
$$
E[y_i] = a + bx_i
$$
$$
var[y_i] = v(E[y_i])
$$
# GLM structure using Gaussian example
## random component
$$
Y_i \sim N(\mu_i, \sigma^2)
$$
$$
Y_i \sim Normal(\mu_i, \sigma^2)
$$
## systematic component
$$
\eta_i = \sum^{p-1}_{n = 0}\beta_jx_{ij}
$$
$$
\mu_i = \beta_0 + \beta x_i
$$
## link
$$
\eta = g(\mu_i)
$$
# GLM structure using binary example
## random
$$
E(Y) = p
$$
## systematic
## link
$$
\eta = g(p) = log(\frac{p_i}{1 - p_i})
$$
$$
log(\frac{p_i}{1 - p_i}) = \beta_0 + \beta_1 x_i
$$
$$
Y_i \sim Binomial(p_i)
$$
$$
\eta = logit(p_i) = log(\frac{p_i}{1 - p_i})
$$
# binomial/bernoulli example
```{r}
set.seed(666)
lizard <- tibble(
pushup = c(rep(1, 30), rep(0, 30)),
distance = c(rnorm(n = 20, mean = 10, sd = 2),
rnorm(n = 20, mean = 20, sd = 2),
rnorm(n = 20, mean = 30, sd = 2))
) %>%
mutate(distance = round(distance, 1))
slice_sample(lizard, n = 10)
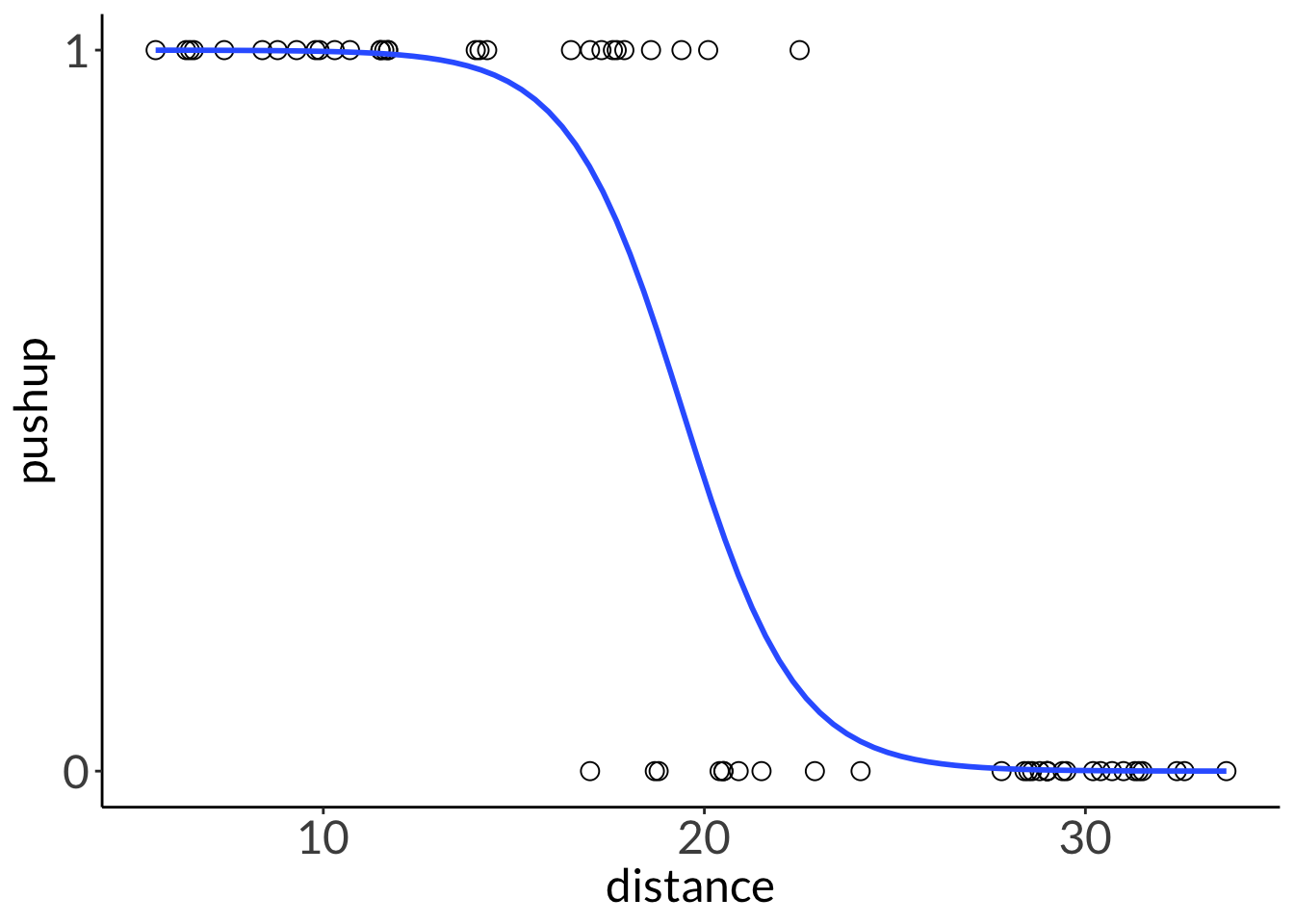
ggplot(lizard,
aes(x = distance,
y = pushup)) +
geom_point(size = 3,
shape = 21) +
scale_y_continuous(limits = c(0, 1),
breaks = c(0, 1)) +
geom_smooth(method = "glm",
method.args = list(family = "binomial"),
se = FALSE,
linewidth = 1)
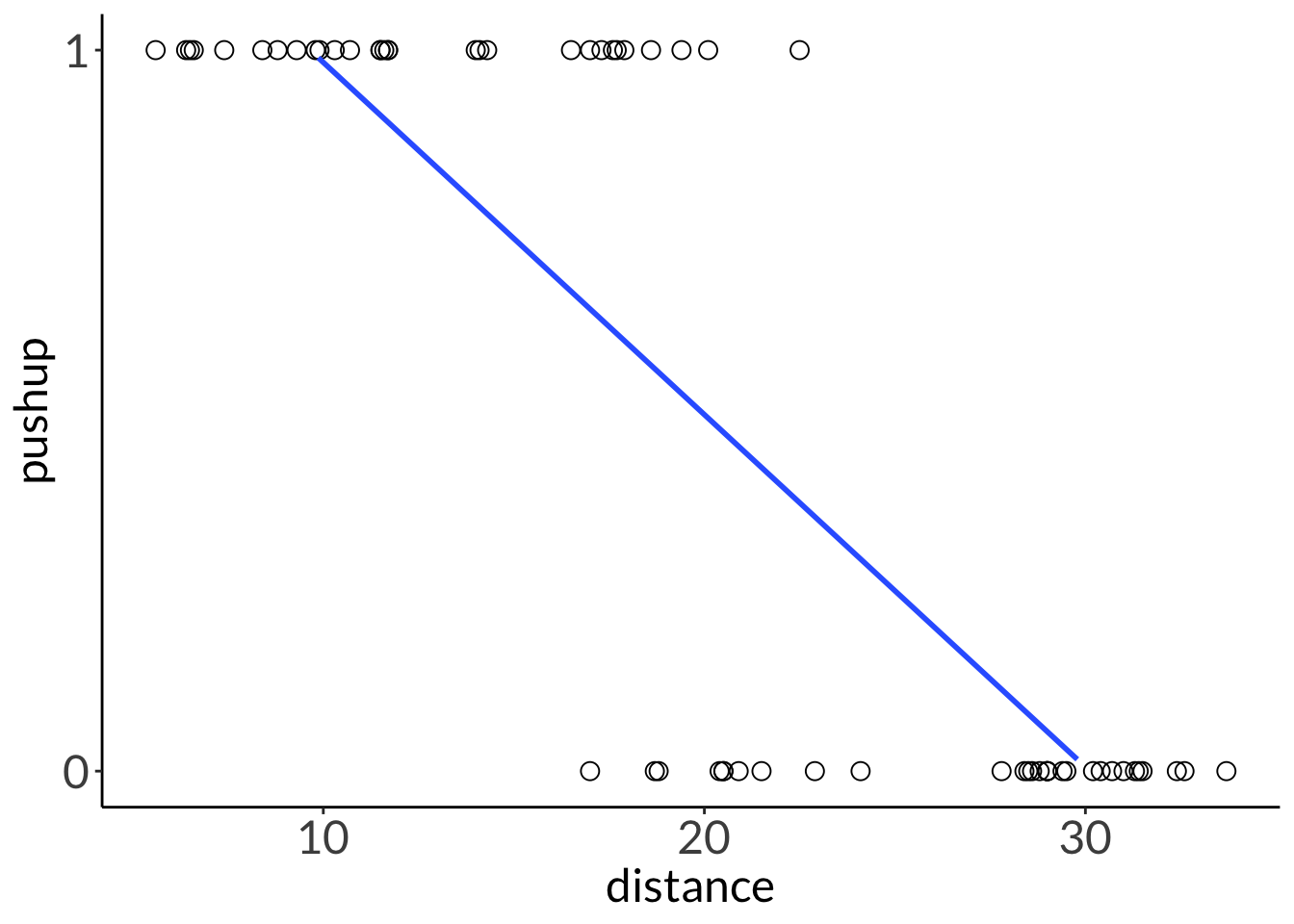
ggplot(lizard,
aes(x = distance,
y = pushup)) +
geom_point(size = 3,
shape = 21) +
scale_y_continuous(limits = c(0, 1),
breaks = c(0, 1)) +
geom_smooth(method = "lm",
se = FALSE,
linewidth = 1)
liz_mod <- glm(pushup ~ distance,
data = lizard,
family = "binomial")
summary(liz_mod)
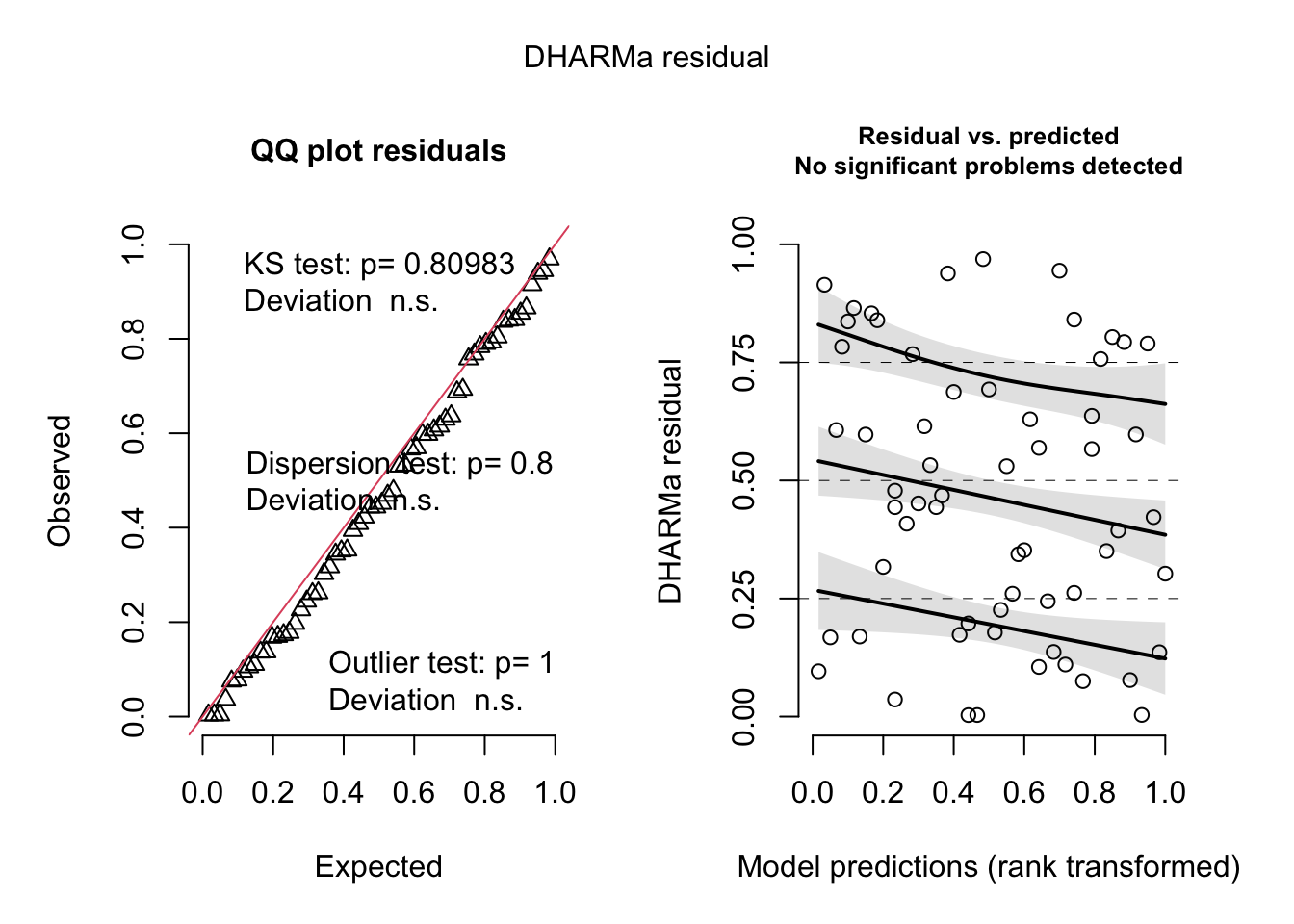
simulateResiduals(liz_mod) %>% plot()
confint(liz_mod)
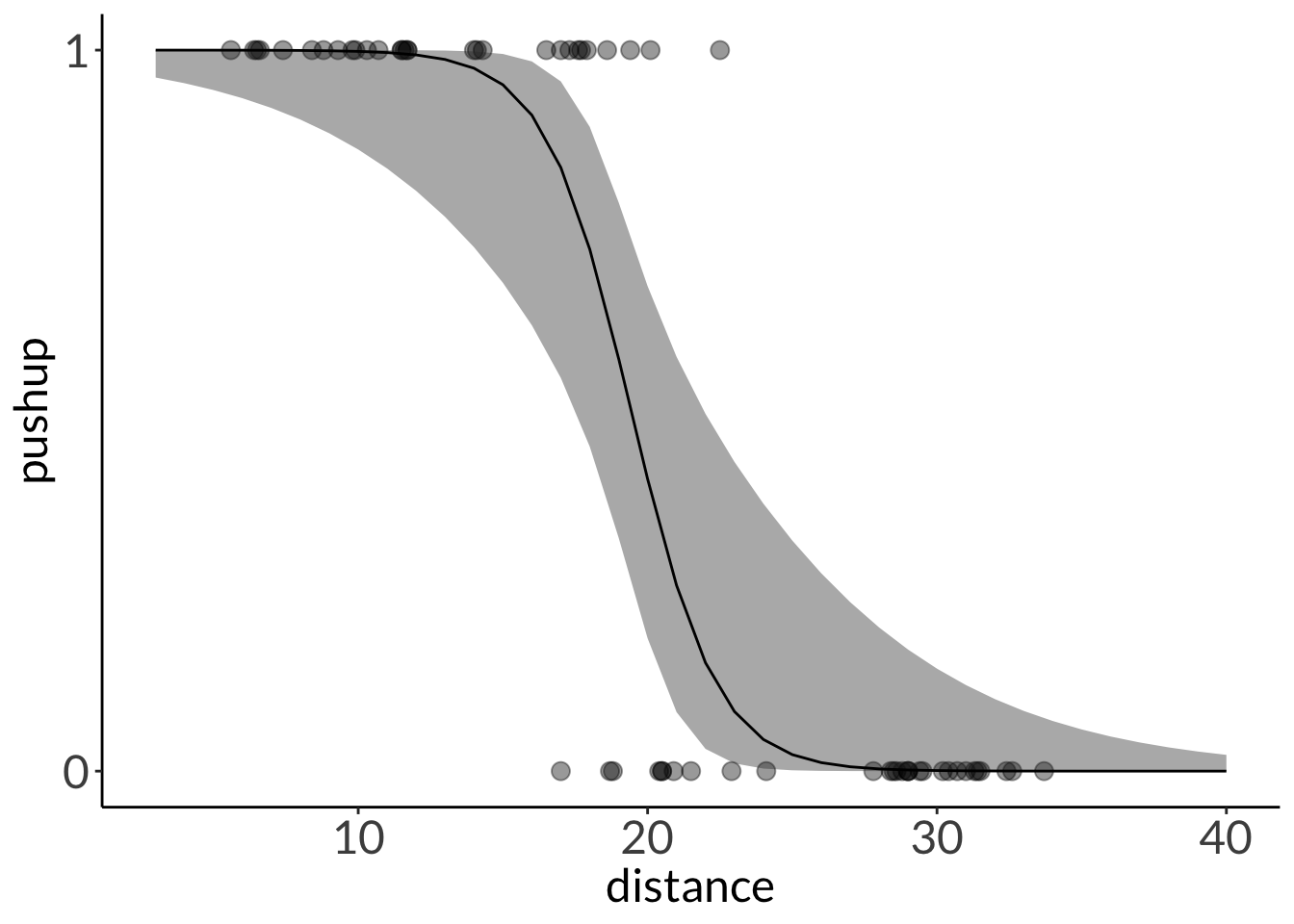
mod_preds <- ggpredict(liz_mod,
terms = "distance [3:40 by = 1]")
ggplot(lizard,
aes(x = distance,
y = pushup)) +
geom_point(size = 3,
alpha = 0.4) +
geom_ribbon(data = mod_preds,
aes(x = x,
y = predicted,
ymin = conf.low,
ymax = conf.high),
alpha = 0.4) +
geom_line(data = mod_preds,
aes(x = x,
y = predicted)) +
scale_y_continuous(limits = c(0, 1),
breaks = c(0, 1))
# what is the probability of a pushup at 20cm?
ggpredict(liz_mod,
terms = "distance [20]")
# what is the probability of a pushup at 10cm?
ggpredict(liz_mod,
terms = "distance [10]")
# what is the probability of a pushup at 30cm?
ggpredict(liz_mod,
terms = "distance [30]")
# what is the probability of a pushup at 20cm?
predict(liz_mod,
newdata = data.frame(distance = 20),
type = "response")
r.squaredLR(liz_mod)
gtsummary::tbl_regression(liz_mod)
gtsummary::tbl_regression(liz_mod,
exponentiate = TRUE)
as_flextable(liz_mod)
```
# negative binomial example
```{r nbinom-fig}
set.seed(666)
nbinom_df <- bind_cols(
size1 = rnbinom(mu = 10, size = 1, n = 100),
size10 = rnbinom(mu = 10, size = 10, n = 100),
size100 = rnbinom(mu = 10, size = 100, n = 100)
)
ggplot(data.frame(x = 1:20), aes(x)) +
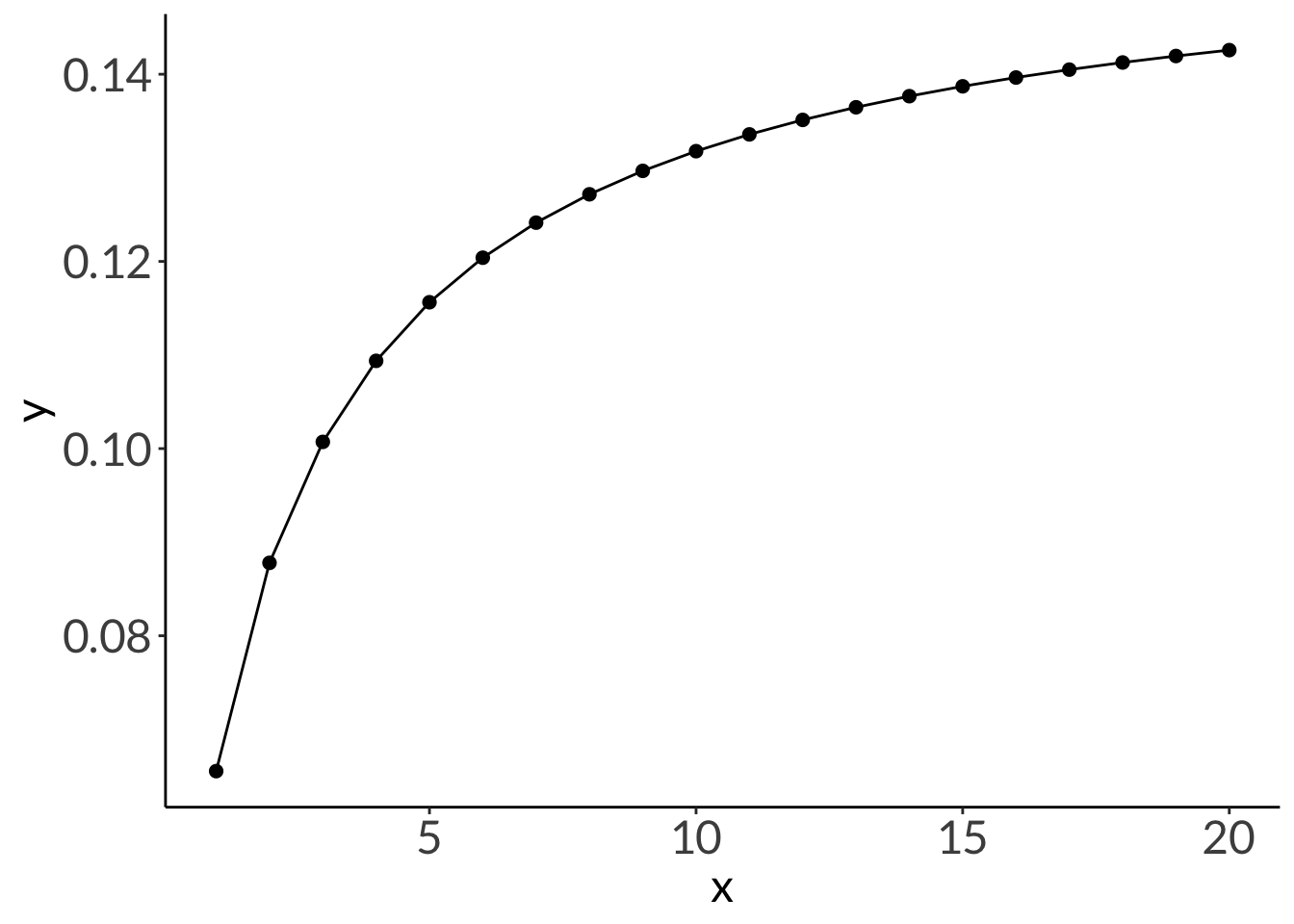
stat_function(geom = "point", n = 20, fun = dnbinom, args = list(mu = 4, x = 5), size = 2) +
stat_function(geom = "line", n = 20, fun = dnbinom, args = list(mu = 4, x = 5))
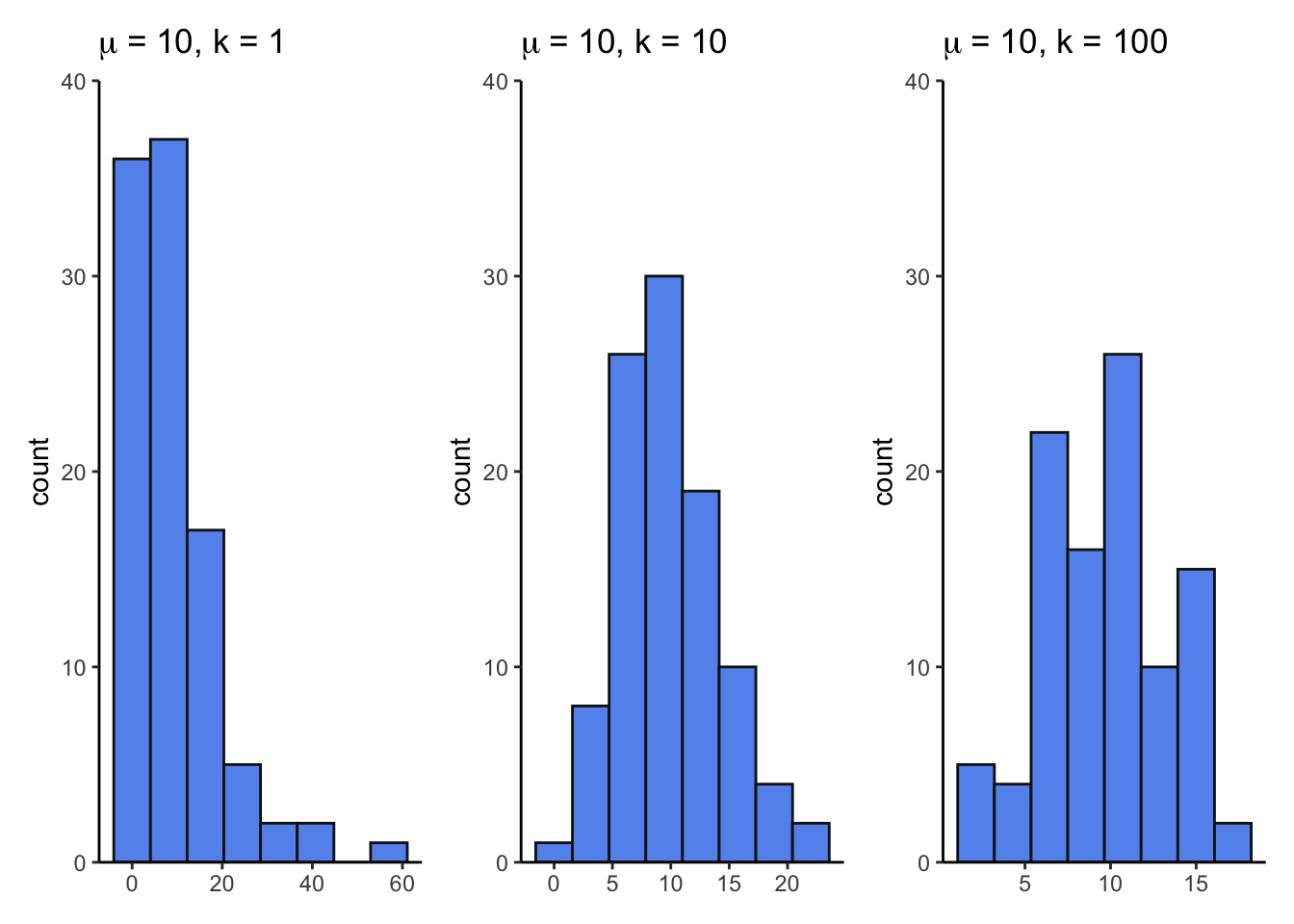
size1 <- ggplot(nbinom_df, aes(x = size1)) +
geom_histogram(bins = 8, fill = "cornflowerblue", color = "grey8") +
scale_y_continuous(expand = c(0, 0), limits = c(0, 40)) +
theme_classic() +
labs(title = expression(mu~"= 10, k = 1")) +
theme(axis.title.x = element_blank())
size10 <- ggplot(nbinom_df, aes(x = size10)) +
geom_histogram(bins = 8, fill = "cornflowerblue", color = "grey8") +
scale_y_continuous(expand = c(0, 0), limits = c(0, 40)) +
theme_classic() +
labs(title = expression(mu~"= 10, k = 10")) +
theme(axis.title.x = element_blank())
size100 <- ggplot(nbinom_df, aes(x = size100)) +
geom_histogram(bins = 8, fill = "cornflowerblue", color = "grey8") +
scale_y_continuous(expand = c(0, 0), limits = c(0, 40)) +
theme_classic() +
labs(title = expression(mu~"= 10, k = 100")) +
theme(axis.title.x = element_blank())
size1 + size10 + size100
```
# poisson example
```{r pois-fig}
set.seed(666)
pois_df <- bind_cols(
lambda1 = rpois(lambda = 1, n = 100),
lambda5 = rpois(lambda = 5, n = 100),
lambda20 = rpois(lambda = 20, n = 100)
)
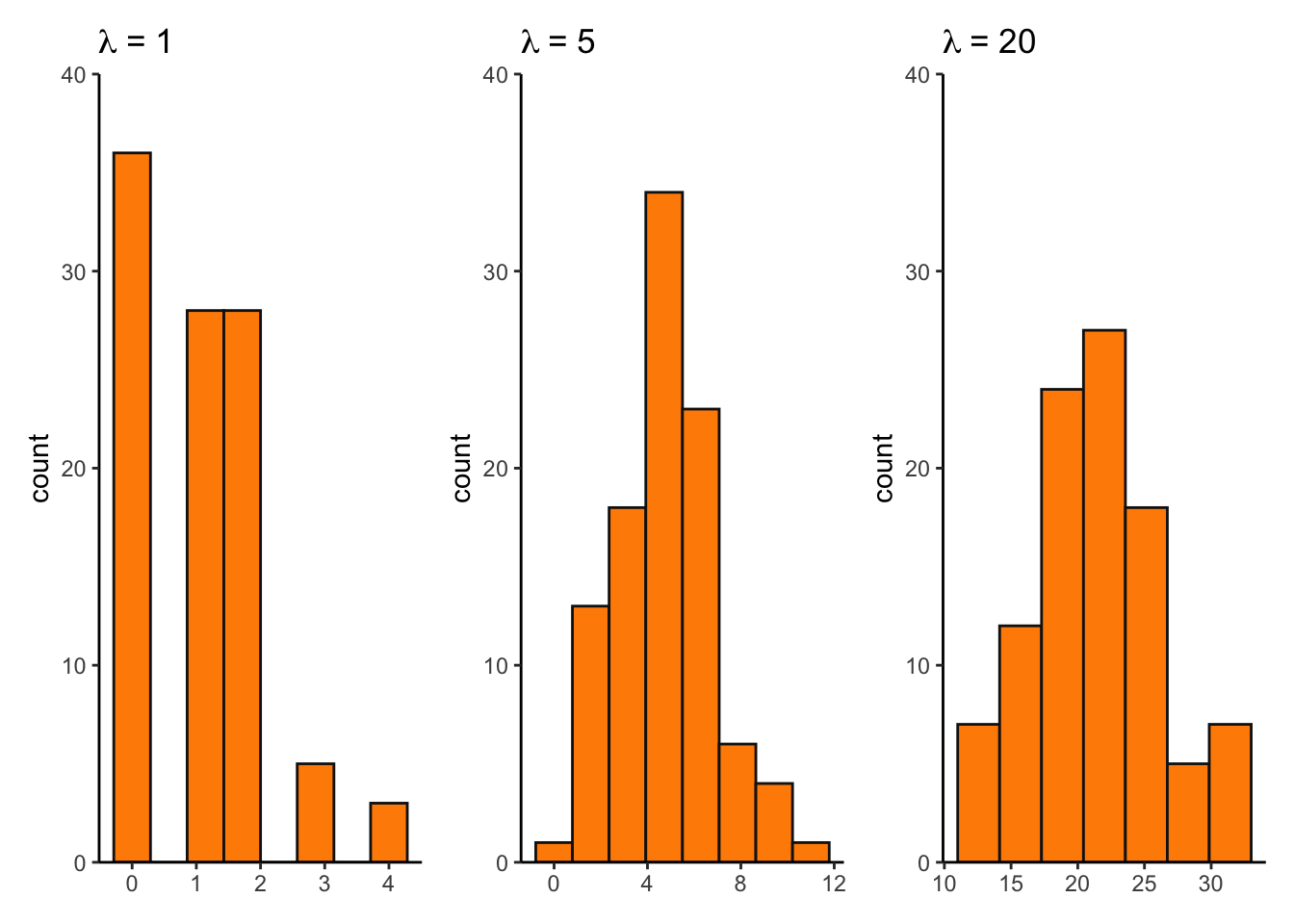
lambda1 <- ggplot(pois_df, aes(x = lambda1)) +
geom_histogram(bins = 8, fill = "darkorange", color = "grey8") +
scale_y_continuous(expand = c(0, 0), limits = c(0, 40)) +
theme_classic() +
labs(title = expression(lambda~"= 1")) +
theme(axis.title.x = element_blank())
lambda5 <- ggplot(pois_df, aes(x = lambda5)) +
geom_histogram(bins = 8, fill = "darkorange", color = "grey8") +
scale_y_continuous(expand = c(0, 0), limits = c(0, 40)) +
theme_classic() +
labs(title = expression(lambda~"= 5")) +
theme(axis.title.x = element_blank())
lambda20 <- ggplot(pois_df, aes(x = lambda20)) +
geom_histogram(bins = 8, fill = "darkorange", color = "grey8") +
scale_y_continuous(expand = c(0, 0), limits = c(0, 40)) +
theme_classic() +
labs(title = expression(lambda~"= 20")) +
theme(axis.title.x = element_blank())
lambda1 + lambda5 + lambda20
```