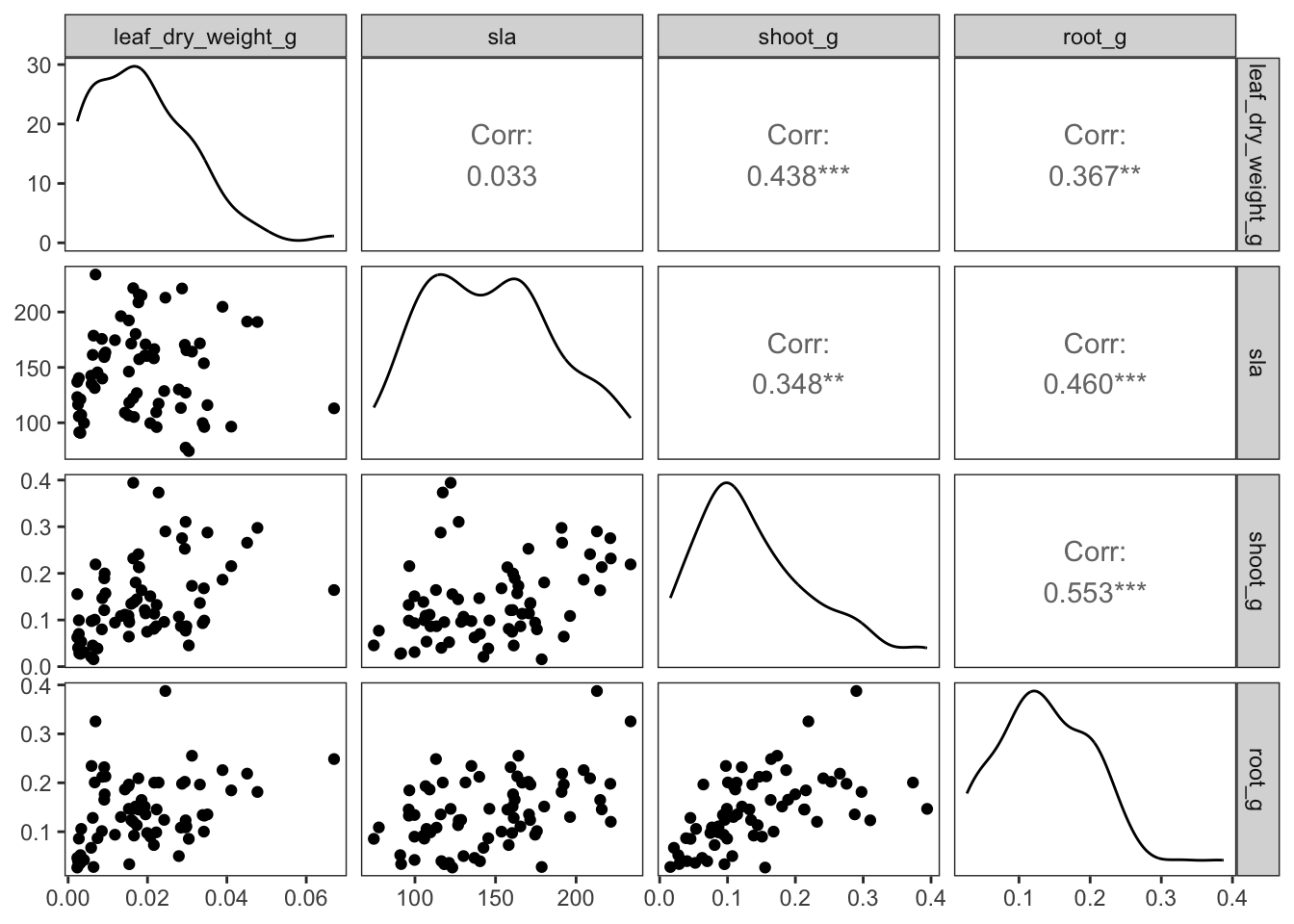
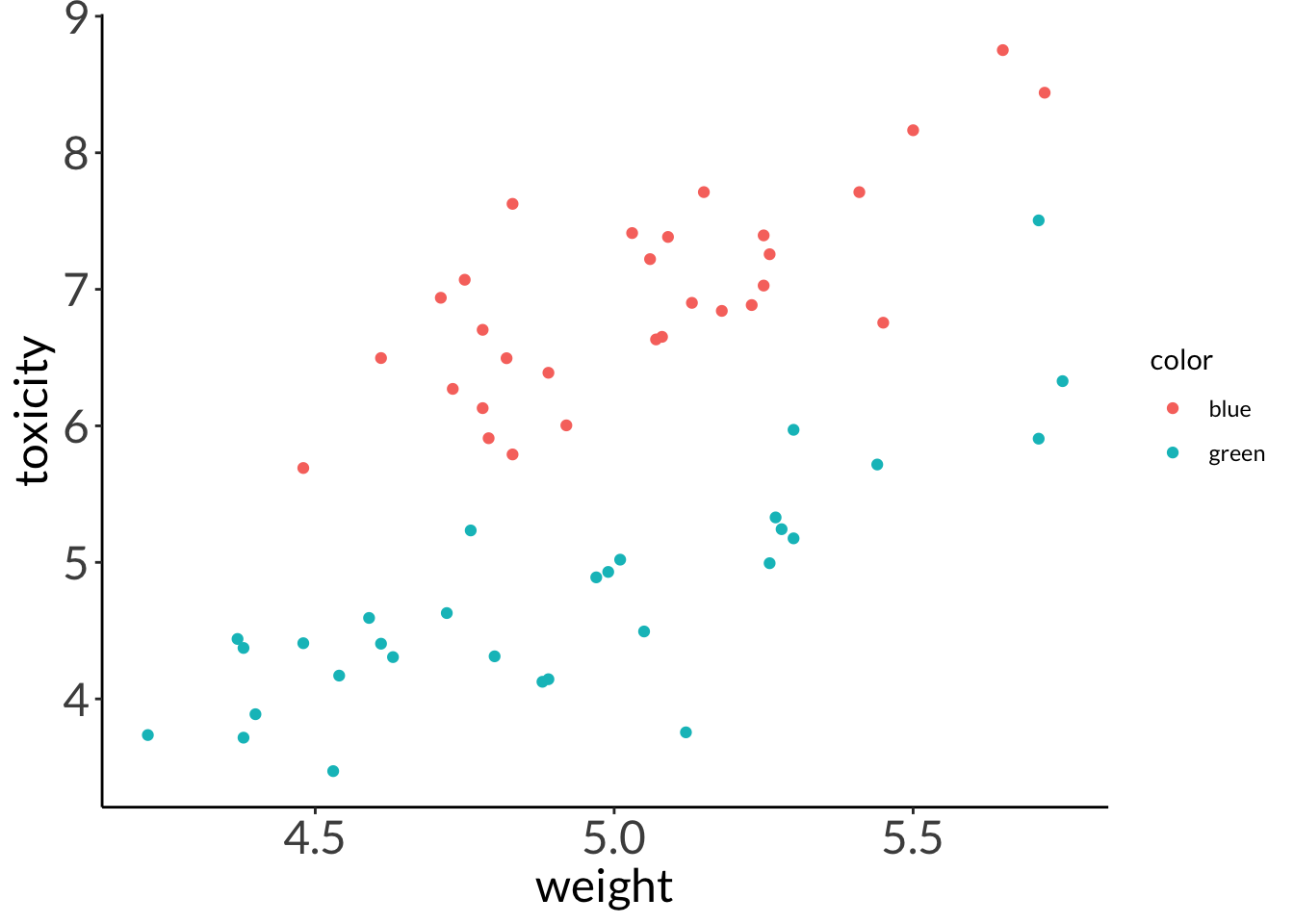
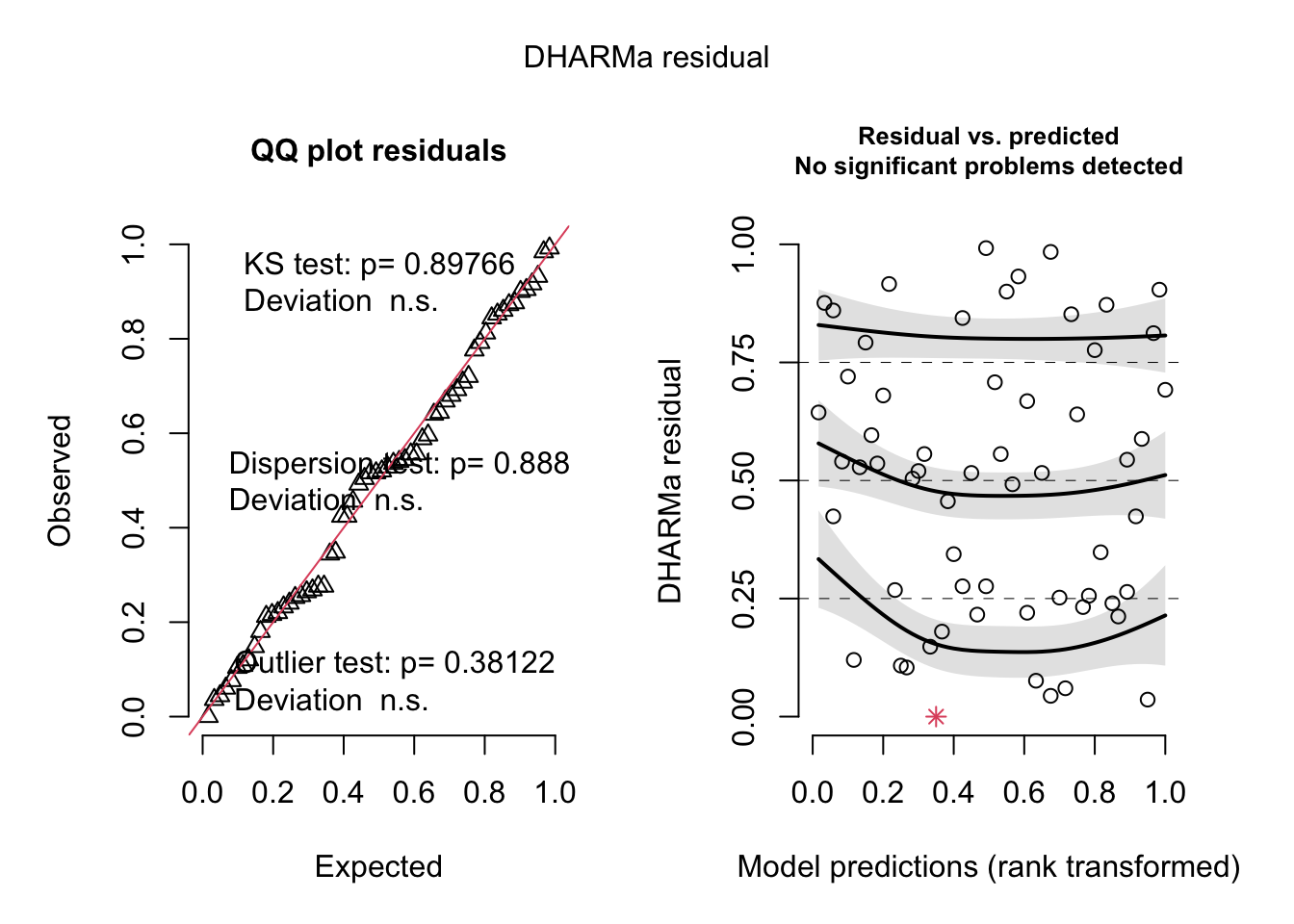
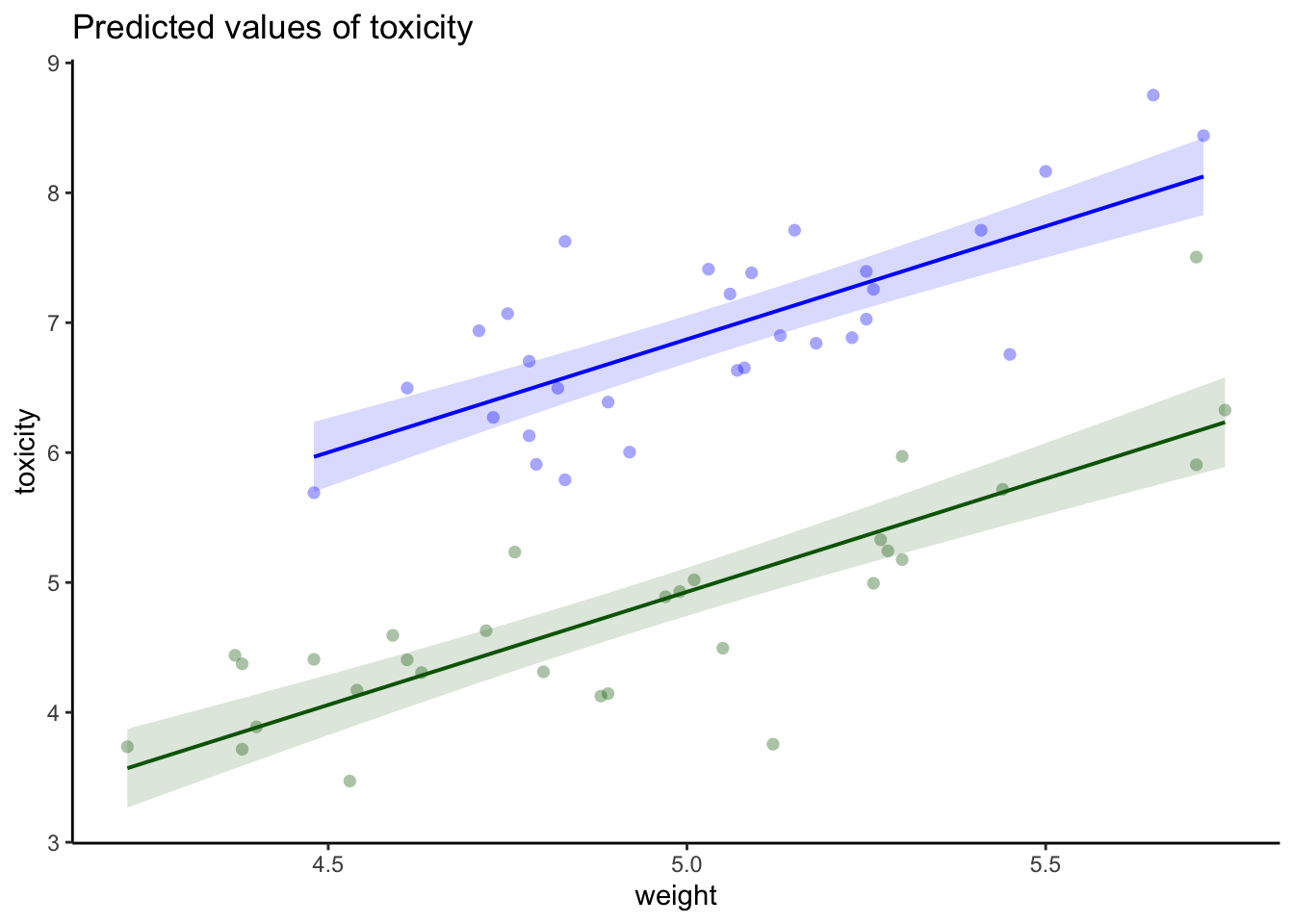
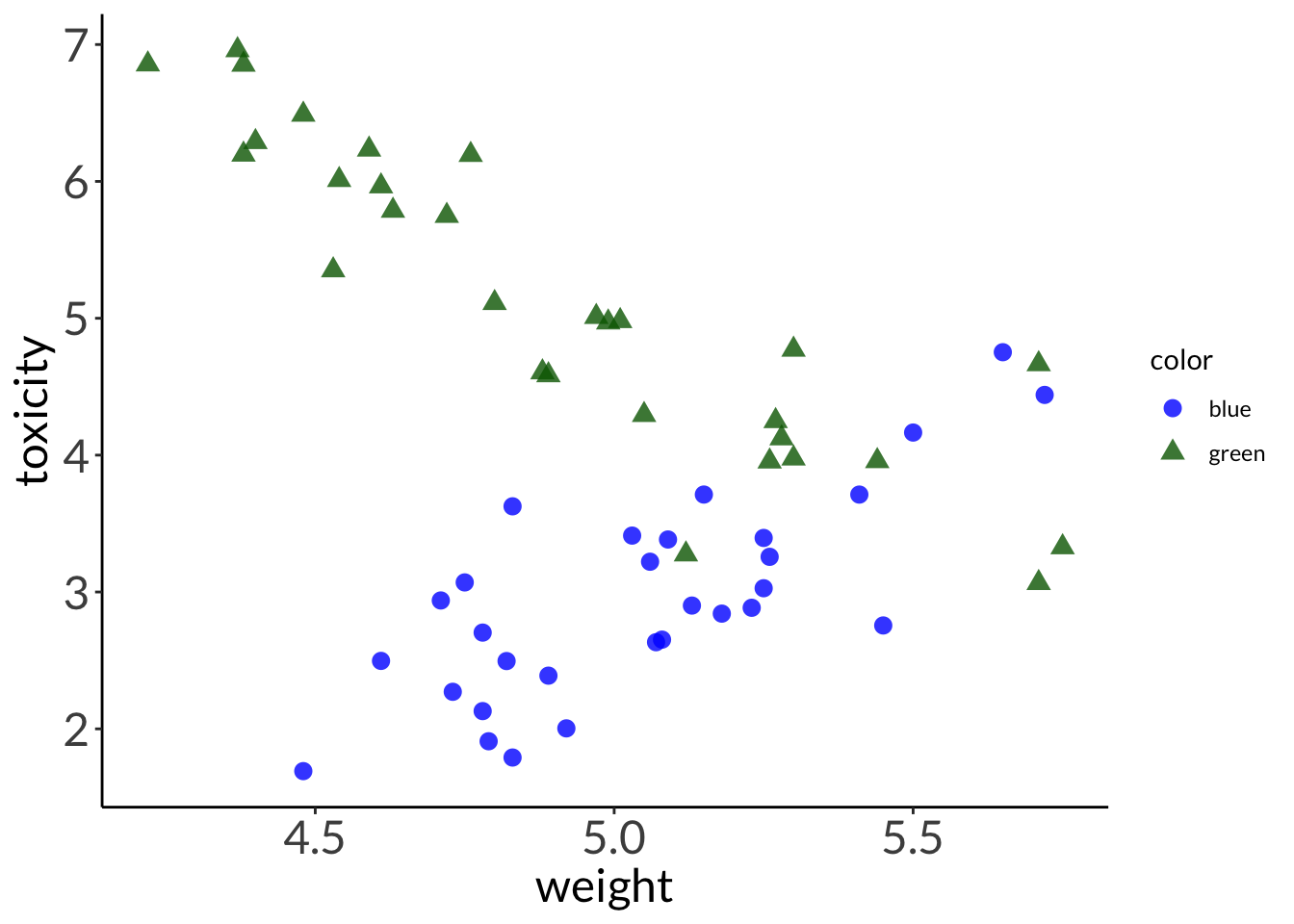
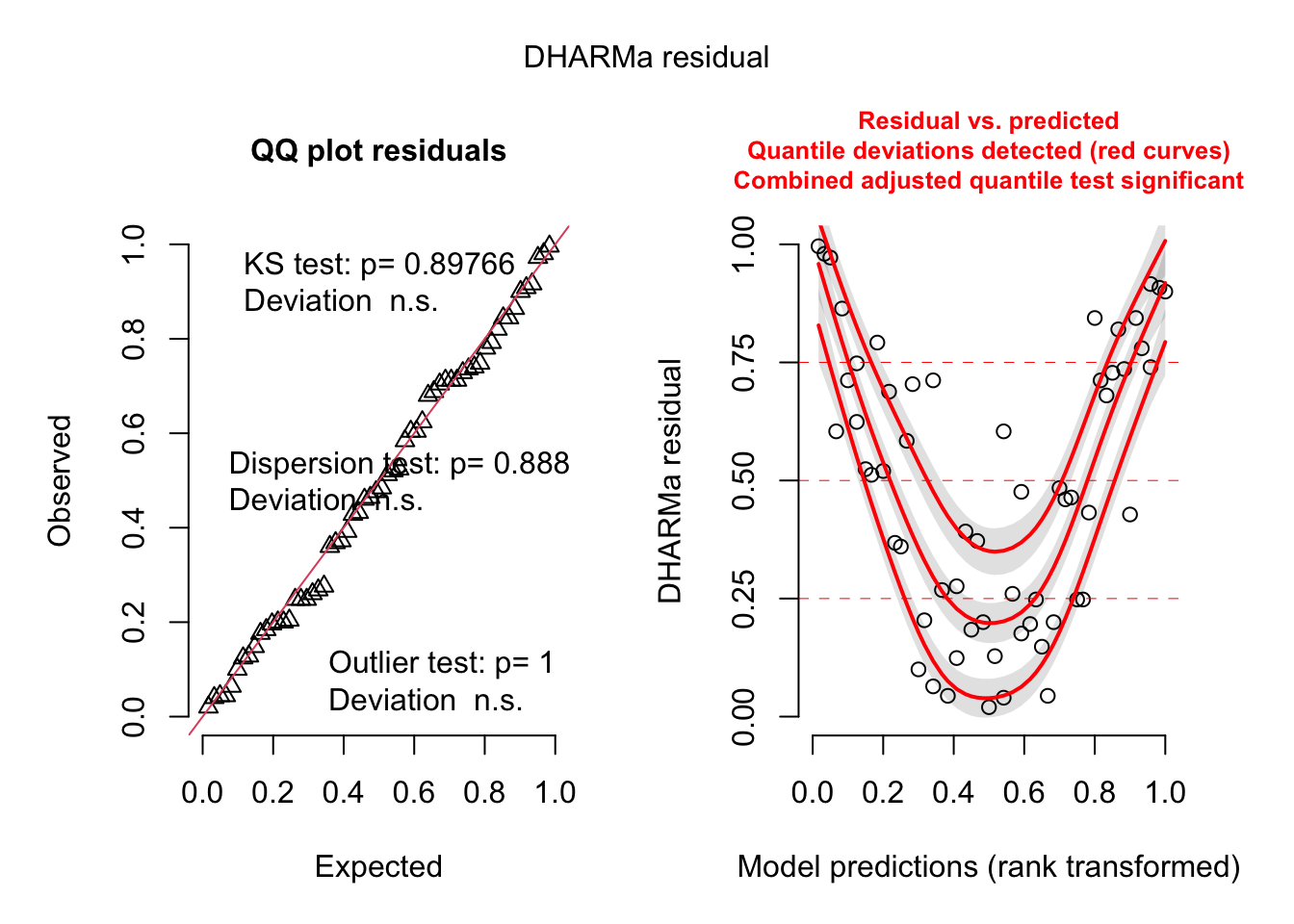
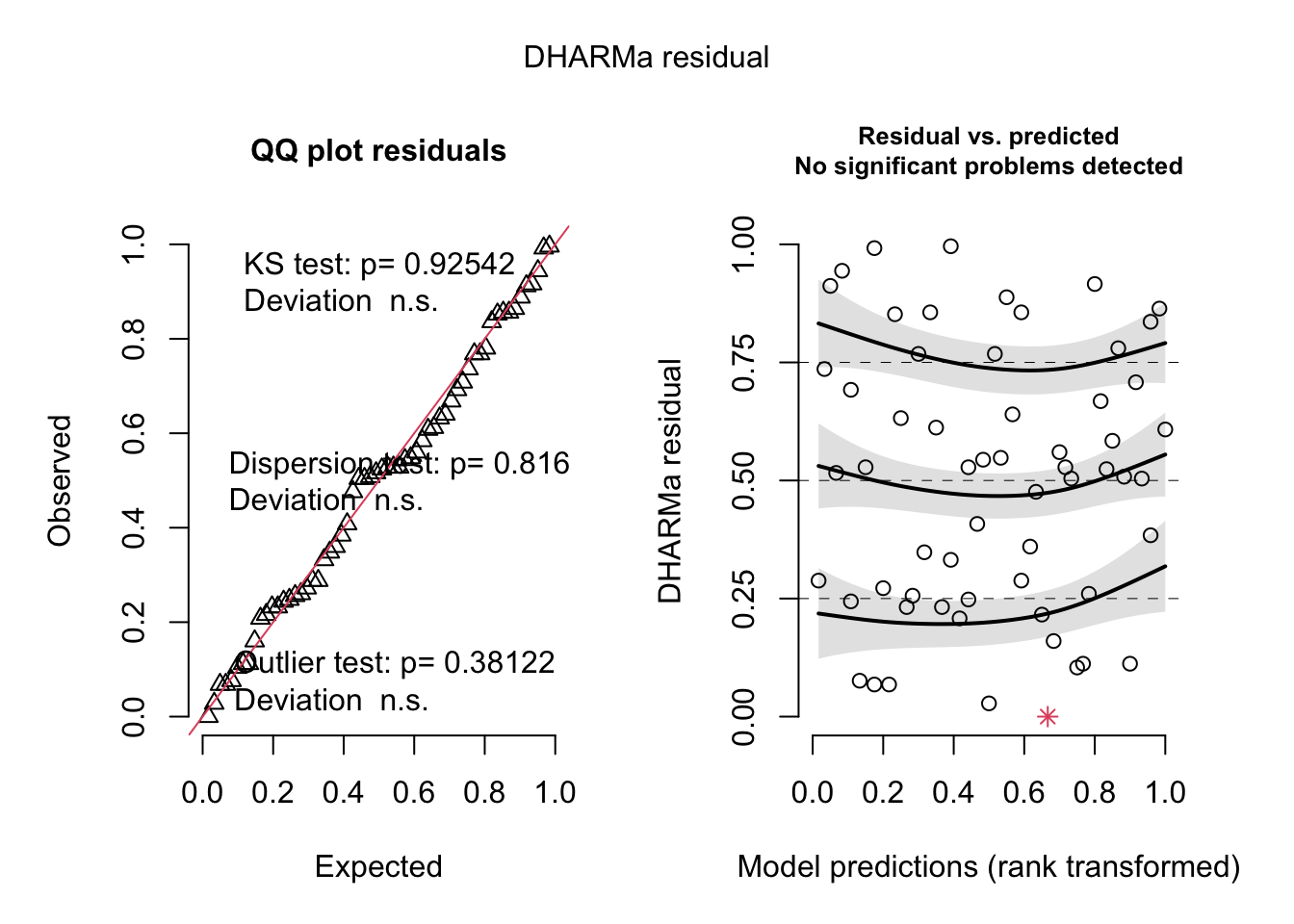
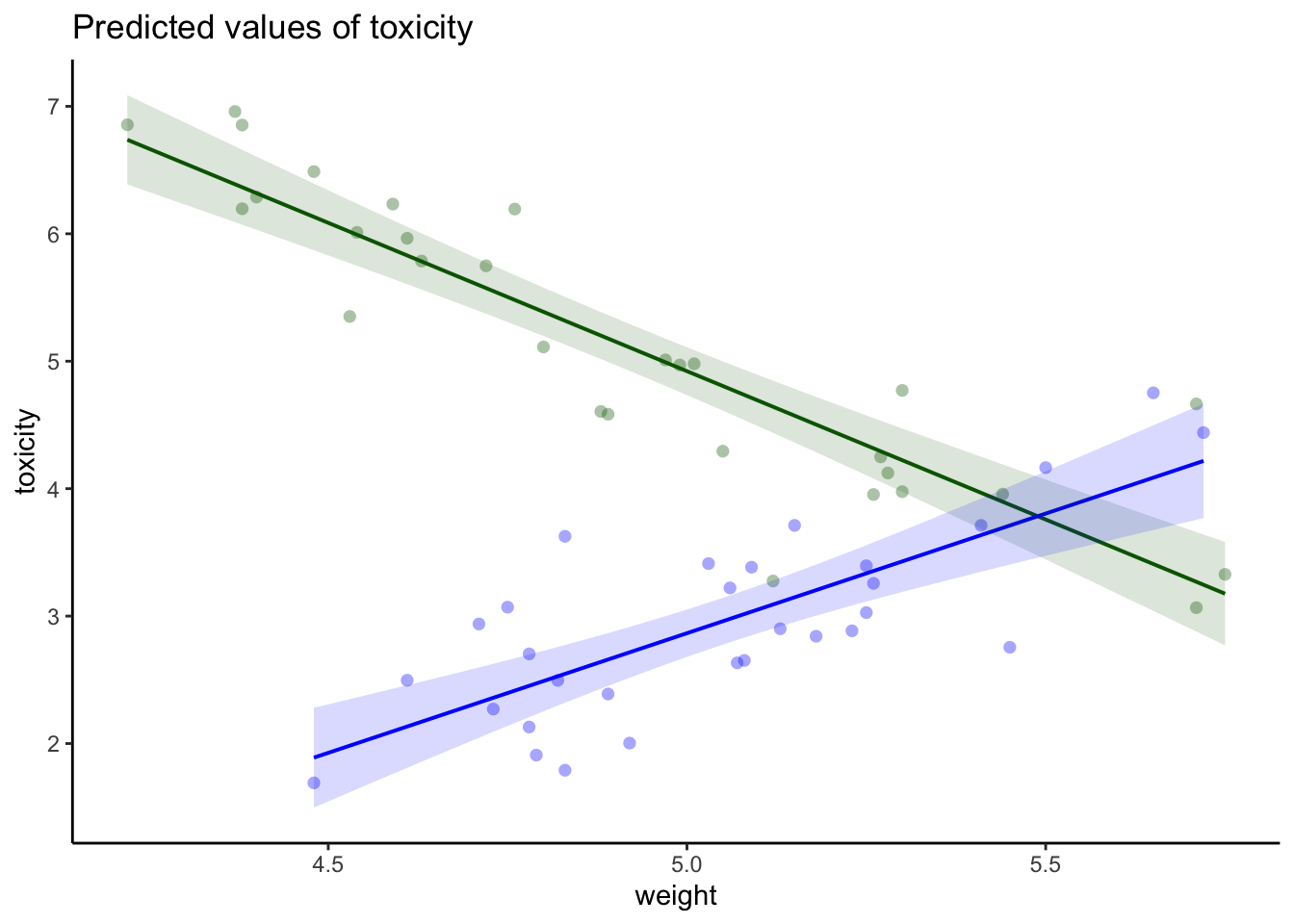
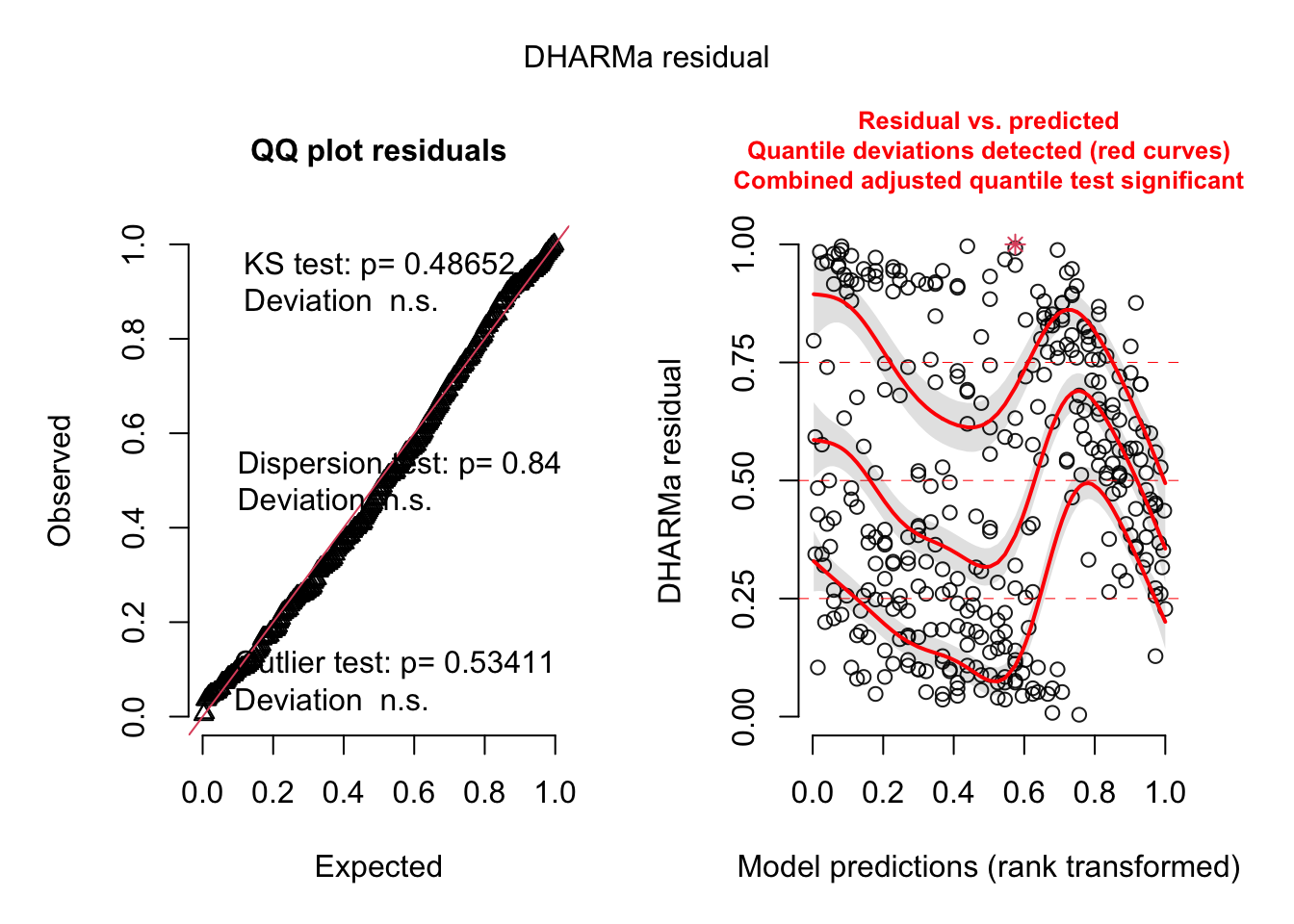
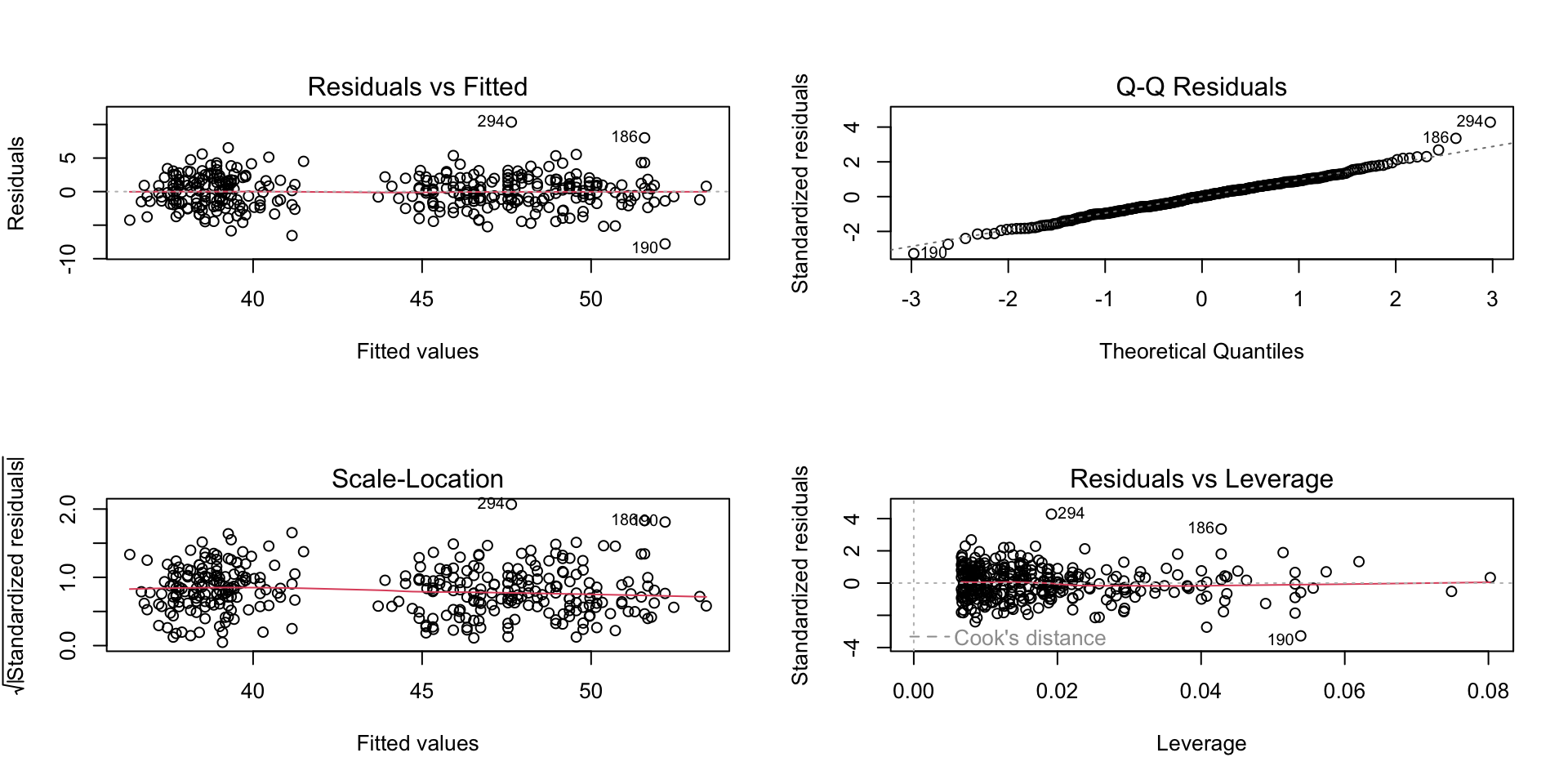
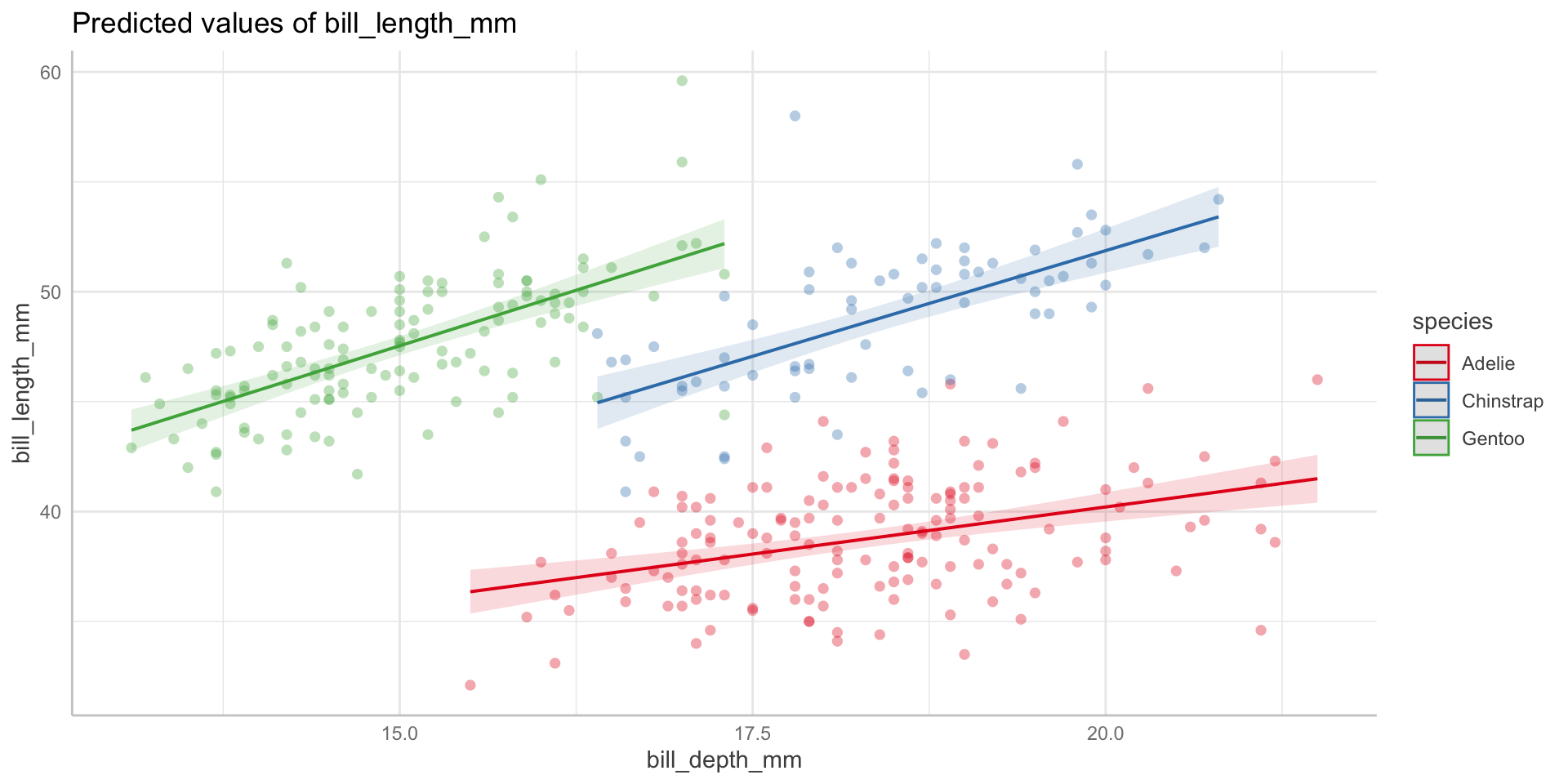
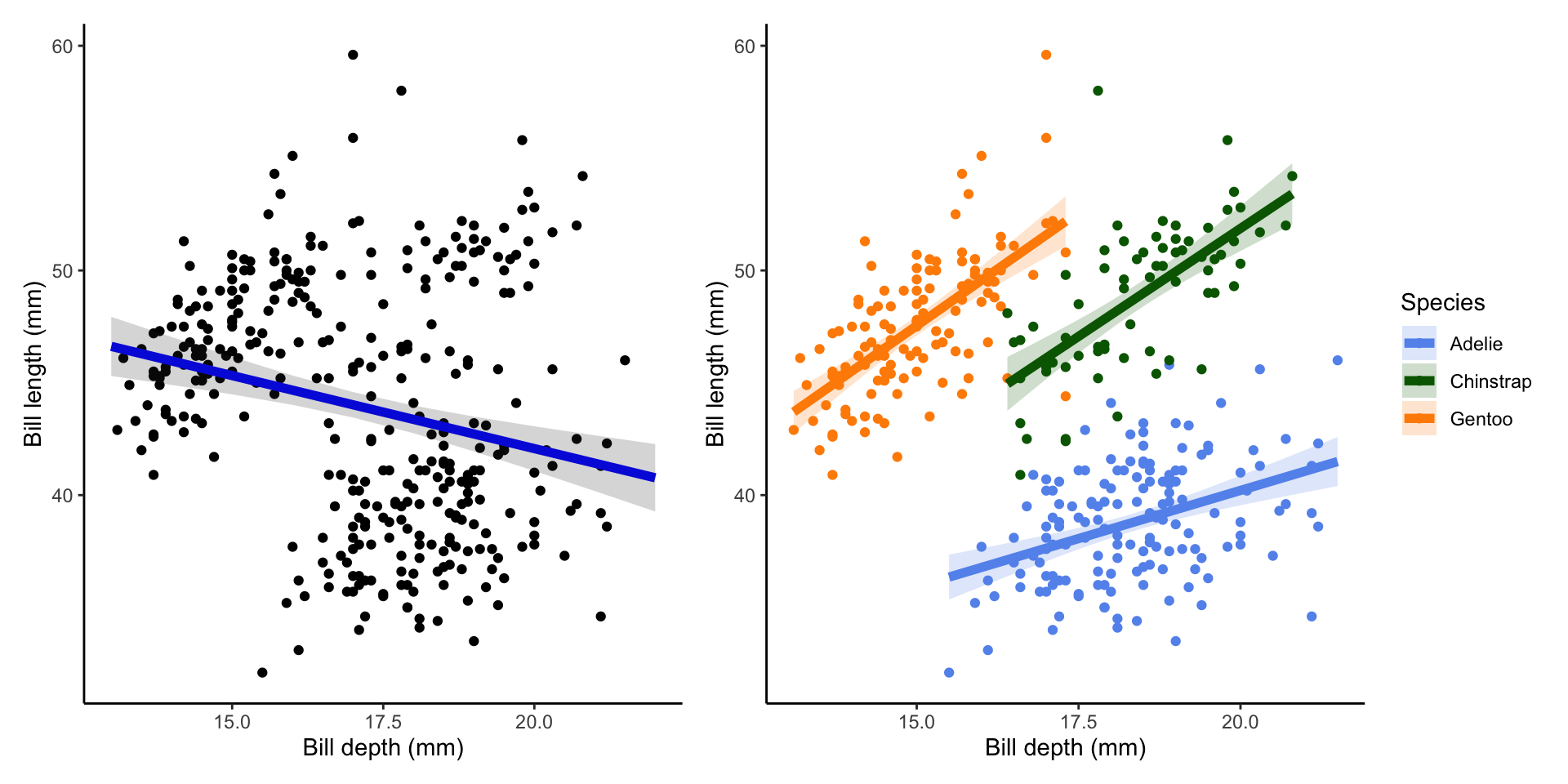
--- title: "Week 9 figures - Lecture 16" editor: source freeze: auto execute: message: false warning: false format: html: code-fold: true author: - name: An Bui url: https://an-bui.com/ affiliation: UC Santa Barbara, Ecology, Evolution, and Marine Biology affiliation-url: https://www.eemb.ucsb.edu/ published-title: "Lecture date" date: 2024-05-29 date-modified: last-modified categories: [multiple linear regression, AIC] citation: url: https://spring-2024.envs-193ds.com/lecture/lecture_week-08.html --- ```{r libraries} # cleaning library(tidyverse) library(readxl) library(here) library(janitor) # visualization theme_set(theme_classic() + theme(panel.grid = element_blank(), axis.text = element_text(size = 18), axis.title = element_text(size = 18), text = element_text(family = "Lato"))) library(patchwork) library(ggeffects) library(flextable) library(GGally) # data library(palmerpenguins) # analysis library(car) library(performance) library(broom) library(DHARMa) library(MuMIn) library(lmtest) library(equatiomatic) ``` ```{r} <- read_xlsx (path = here ("data" , "Valliere_etal_EcoApps_Data.xlsx" ),sheet = "First Harvest" )# cleaning <- drought_exp %>% clean_names () %>% # nicer column names mutate (species_name = case_when ( # adding column with species scientific names == "ENCCAL" ~ "Encelia californica" , # bush sunflower == "ESCCAL" ~ "Eschscholzia californica" , # California poppy == "PENCEN" ~ "Penstemon centranthifolius" , # Scarlet bugler == "GRICAM" ~ "Grindelia camporum" , # great valley gumweed == "SALLEU" ~ "Salvia leucophylla" , # Purple sage == "STIPUL" ~ "Nasella pulchra" , # Purple needlegrass == "LOTSCO" ~ "Acmispon glaber" # deerweed %>% relocate (species_name, .after = species) %>% # moving species_name column after species mutate (water_treatment = case_when ( # adding column with full treatment names == "WW" ~ "Well watered" ,== "DS" ~ "Drought stressed" %>% relocate (water_treatment, .after = water) # moving water_treatment column after water ``` # math ## variance inflation factor ```{r} ggpairs (drought_exp_clean, # data frame columns = c ("leaf_dry_weight_g" , # columns to visualize "sla" , "shoot_g" , "root_g" ), upper = list (method = "pearson" )) + # calculating Pearson correlation coefficient theme_bw () + # cleaner theme theme (panel.grid = element_blank ()) # getting rid of gridlines ``` # VIF ```{r} <- lm (total_g ~ leaf_dry_weight_g + sla + root_g + shoot_g,data = drought_exp_clean)vif (plant_model_all)<- lm (total_g ~ leaf_dry_weight_g + sla + r_s,data = drought_exp_clean)vif (plant_model_simple)<- lm (total_g ~ sla + water_treatment + species,data = drought_exp_clean)vif (model1)``` # interaction terms ## no interaction ```{r} <- 30 set.seed (666 )<- tibble (weight = c (round (rnorm (n = frog_num, mean = 5 , sd = 0.35 ), 2 ),round (rnorm (n = frog_num, mean = 5 , sd = 0.35 ), 2 )color = c (rep ("green" , frog_num),rep ("blue" , frog_num)%>% mutate (toxicity = case_when (== "green" ~ round (rnorm (n = frog_num, mean = 2 , sd = 0.1 ), 2 )* weight - 5 ,== "blue" ~ round (rnorm (n = frog_num, mean = 2 , sd = 0.1 ), 2 )* weight - 3 ggplot (data = frogs,aes (x = weight,y = toxicity,color = color)) + geom_point ()<- lm (toxicity ~ weight + color,data = frogs)simulateResiduals (frog_mod) %>% plot ()summary (frog_mod)ggpredict (frog_mod,terms = c ("weight [all]" ,"color" )) %>% plot (show_data = TRUE , limit_range = TRUE ) + scale_color_manual (values = c ("green" = "darkgreen" , "blue" = "blue" )) + scale_fill_manual (values = c ("green" = "darkgreen" , "blue" = "blue" )) + theme_classic () + theme (legend.position = "none" )``` ## with interaction ```{r} set.seed (666 )<- tibble (weight = c (round (rnorm (n = frog_num, mean = 5 , sd = 0.35 ), 2 ),round (rnorm (n = frog_num, mean = 5 , sd = 0.35 ), 2 )color = c (rep ("green" , frog_num),rep ("blue" , frog_num)%>% mutate (toxicity = case_when (== "green" ~ round (rnorm (n = frog_num, mean = - 2 , sd = 0.1 ), 2 )* weight + 15 ,== "blue" ~ round (rnorm (n = frog_num, mean = 2 , sd = 0.1 ), 2 )* weight - 7 ggplot (data = frogs,aes (x = weight,y = toxicity,color = color,shape = color)) + geom_point (size = 3 ,alpha = 0.8 ) + # scale_x_continuous(limits = c(0, 7)) + scale_color_manual (values = c ("green" = "darkgreen" , "blue" = "blue" )) <- lm (toxicity ~ weight + color, # no interaction data = frogs)<- lm (toxicity ~ weight * color, # interaction data = frogs)simulateResiduals (frog_mod1) %>% plot () # no interaction simulateResiduals (frog_mod2) %>% plot () # interaction summary (frog_mod1) # no interaction summary (frog_mod2) # interaction model.sel (frog_mod1, # no interaction # interaction ggpredict (frog_mod2,terms = c ("weight [all]" ,"color" )) %>% plot (show_data = TRUE , limit_range = TRUE ) + scale_color_manual (values = c ("green" = "darkgreen" , "blue" = "blue" )) + scale_fill_manual (values = c ("green" = "darkgreen" , "blue" = "blue" )) + theme_classic () + theme (legend.position = "none" )``` ```{r} <- ggpredict (frog_mod2,terms = c ("weight [all]" ,"color" )) %>% group_by (group) %>% filter (predicted %in% c (max (predicted), min (predicted))) %>% arrange (group)# solve for slopes <- (pluck (predictions, 2 , 2 ) - pluck (predictions, 2 , 1 ))/ (pluck (predictions, 1 , 2 ) - pluck (predictions, 1 , 1 ))<- (pluck (predictions, 2 , 4 ) - pluck (predictions, 2 , 3 ))/ (pluck (predictions, 1 , 4 ) - pluck (predictions, 1 , 3 ))# solve for intercepts <- pluck (predictions, 2 , 2 ) - (green_slope* pluck (predictions, 1 , 2 ))<- pluck (predictions, 2 , 4 ) - (blue_slope* pluck (predictions, 1 , 4 ))ggpredict (frog_mod2,terms = c ("weight [5]" ,"color" ))* 5 + green_intercept* 5 + blue_intercept``` # simpson's paradox ```{r} <- lm (bill_length_mm ~ bill_depth_mm, data = penguins)simulateResiduals (bill_model) %>% plot ()summary (bill_model)confint (bill_model)<- ggpredict (bill_model, terms = "bill_depth_mm" )<- ggplot (penguins, aes (x = bill_depth_mm, y = bill_length_mm)) + geom_point () + geom_line (data = bill_model_preds, aes (x = x, y = predicted), color = "blue" , linewidth = 2 ) + geom_ribbon (data = bill_model_preds, aes (x = x, y = predicted, ymin = conf.low, ymax = conf.high), alpha = 0.2 ) + theme_classic () + labs (x = "Bill depth (mm)" , y = "Bill length (mm)" )``` ```{r} #| fig-width: 10 <- lm (bill_length_mm ~ bill_depth_mm* species, data = penguins)par (mfrow = c (2 , 2 ))plot (bill_model2)summary (bill_model2)lrtest (bill_model2)anova (bill_model2)waldtest (bill_model2)ggpredict (bill_model2, terms = c ("bill_depth_mm [all]" , "species" )) %>% plot (show_data = TRUE , limit_range = TRUE )%>% group_by (species) %>% summarize (min = min (bill_depth_mm, na.rm = TRUE ),max = max (bill_depth_mm, na.rm = TRUE ))<- ggpredict (bill_model2, terms = c ("bill_depth_mm [13.1:21.5 by = 0.1]" , "species" )) %>% as_tibble () %>% rename (species = group) %>% mutate (keep = case_when (== "Adelie" & between (x, 15.5 , 21.5 ) ~ "keep" ,== "Chinstrap" & between (x, 16.4 , 20.8 ) ~ "keep" ,== "Gentoo" & between (x, 13.1 , 17.3 ) ~ "keep" %>% drop_na (keep) %>% select (- keep)<- ggplot (penguins, aes (x = bill_depth_mm, y = bill_length_mm)) + geom_point (aes (color = species)) + geom_line (data = bill_model2_preds, aes (x = x, y = predicted, color = species), linewidth = 2 ) + geom_ribbon (data = bill_model2_preds, aes (x = x, y = predicted, ymin = conf.low, ymax = conf.high, fill = species), alpha = 0.2 ) + scale_color_manual (values = c ("cornflowerblue" , "darkgreen" , "darkorange" )) + scale_fill_manual (values = c ("cornflowerblue" , "darkgreen" , "darkorange" )) + theme_classic () + labs (x = "Bill depth (mm)" , y = "Bill length (mm)" ,color = "Species" , fill = "Species" ) + model2_plotextract_eq (bill_model)``` \_ length\_ mm} = \alpha + \beta_{1}(\operatorname{bill\_ depth\_ mm}) + \beta_{2}(\operatorname{species}_{\operatorname{Chinstrap}}) + \beta_{3}(\operatorname{species}_{\operatorname{Gentoo}}) + \beta_{4}(\operatorname{bill\_depth\_mm} \times \operatorname{species}_{\operatorname{Chinstrap}}) + \beta_{5}(\operatorname{bill\_depth\_mm} \times \operatorname{species}_{\operatorname{Gentoo}}) + \epsilon\_ length\_ mm} = \alpha + \beta_{1}(\operatorname{bill\_ depth\_ mm}) + \epsilon